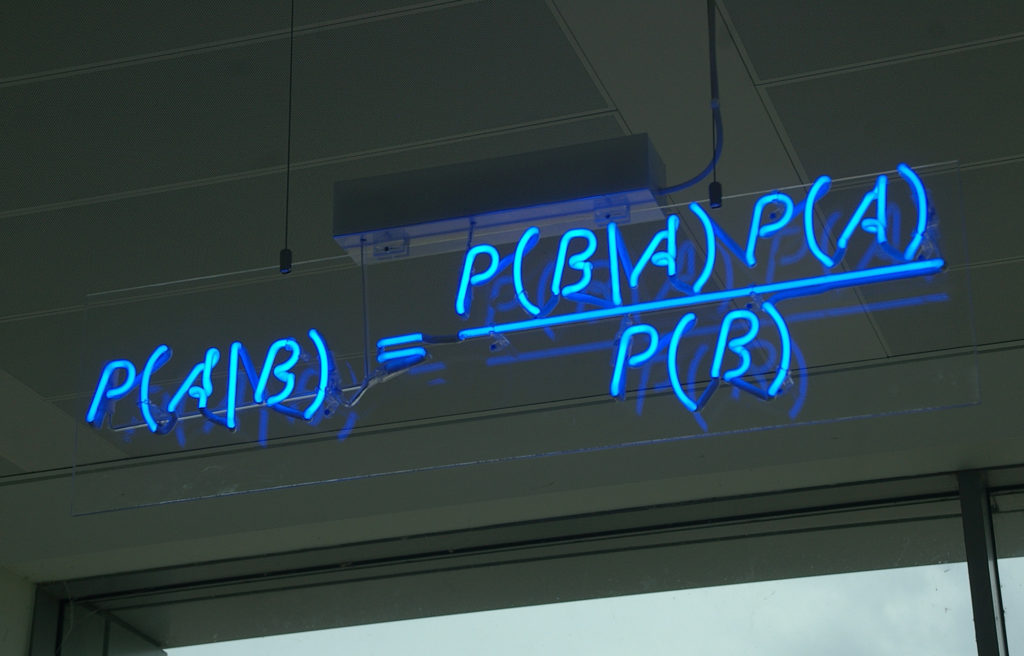Van het Regeerprogramma en de Miljoenennota van het huidige kabinet kun je vinden wat je wilt. Dat doe ik dan ook. Bezuinigingen op zorg, onderwijs, cultuur en wetenschap zijn maatregelen waar ik me meestal niet achter schaar. Over één onderwerp verwacht je dat ik, als onderzoeker in de data science en AI in de populatiegerichte zorg, wel positief zou zijn: meer geld voor AI in de zorg, zodat de administratieve last met de helft zou kunnen afnemen. Maar helaas, mevrouw Agema, het tegendeel is waar. Ik geloof er niet in, en zeker niet op de korte termijn.
De administratieve last in de zorg zal niet door AI worden opgelost, en zeker niet op de korte termijn.
De techniek is niet het probleem. We zijn inderdaad in staat om een stuk software met de arts en patiënt mee te laten luisteren, of wellicht zelfs bij een behandeling mee te laten kijken, en deze te vertalen naar electronische patiëntendossiers of andere vormen van verslaglegging. Op dit moment zorgt dit nog voor administartieve druk. Zelf ben ik ook actief betrokken naar onderzoek om deze technieken verder te verbeteren, al zijn ze naar mijn mening ook al goed genoeg en klaar voor gebruik in de praktijk. Dat gebeurt ook: in een flink aantal Nederlandse ziekenhuizen en andere zorginstellingen worden AI modellen gebruikt zoals hierboven beschreven.
Het kúnnen is dus niet waar de schoen wringt. Vertrouwen in de systemen die hiervoor moeten worden opgetuigd en de benodigde (IT) organisatie áchter die systemen zijn echter wel het probleem. Foutjes zijn snel gemaakt, maar het blijkt in de praktijk dat het veel makkelijker te behapstukken is wanneer foutjes in medische dossiers of behandelplannen zijn geïntroduceerd door menselijk handelen, dan wanneer deze foutjes door toedoen van een computer of robot binnensluipen. We zijn voor mensen vergevingsgezind, voor een AI niet. Gevolg daarvan is dat controle van het door AI gegenereerde verslag een belangrijke nadruk zal krijgen, en dat eventuele missers die er toch doorheen sluipen een veel grotere nasleep zullen krijgen dan de menselijke foutjes die nu optreden. En wat als een AI een diagnose stelt of conlusie trekt die de zorgverlener niet kan duiden, hoe gaat een professional daar dan mee om? In hoeverre moeten professionals dit klakkeloos overnemen, en in hoeverre moeten zij besluiten dit “menings-” of “interpretatieverschil” uit te zoeken? Ineens kost de AI een berg extra tijd.
Ook wet- en regelgeving zijn nu nog niet klaar voor het laten overnemen van administratie door een AI. Als verkeerde verslaglegging resulteert in handelingen die niet goed uitpakken (verkeerde diagnoses, verwijzingen, voorschriften, behandelingen etc.), waar ligt dan de verantwoordelijkheid? Zolang deze bij de mens ligt die anders de verslaglegging had gedaan, is de tijdwinst door een AI vele male lager dan je op eerste gezicht zou denken. Daar komen nog de gevaarlijke missers bovenop die we onlangs hebben langs zien komen: roep de hulp van een AI in voor sommige van je praktische taken, en voor je het weet ligt zeer gevoelige persoonsinformatie op een stoep in Silicon Valley.
Administratieve last zal zich verplaatsen van typen, naar controle en naar het oplossen van mismatches tussen mens en technologie.
Tenslotte is er nog het aspect van het ontwikkelen en onderhouden van software. Software moet een behoorlijk vertrouwen krijgen van maatschappij, arts én patiënt, zodat we deze zonder al te veel handmatige controle en risico op foutjes kunnen inzetten. Dit vergt een (zorgvuldig en gecontroleerd!) ontwikkelproces. Ontwikkeling is duur. Daarna moet de software worden ingezet, en dus onderhouden op een zeer breed verspreid netwerk van computers bij zorgveleners in het ziekenhuis, maar ook in de huisartsenpraktijk en alle andere zorgveleners in de eerste, tweede en derde lijn. Als het dan al in staat blijkt administratieve last weg te nemen bij de zorgprofessional (wat een mooie winst zou zijn), dan nog besparen we niks: het geld verplaatst dan naar de ontwikkeling en het onderhoud van software. Ik denk dat we in de afgelopen jaren genoeg voorbeelden hebben gezien van hoe duur dat uiteindelijk blijkt. Ook een financiële verlichting van de zorg lijkt me op korte termijn niet realistisch.
Kosten zullen verplaatsen van salaris van zorgprofessionals, naar salaris van IT-personeel en naar de rekening van software-ontwikkelende bedrijven met een winstoogmerk.
Wat veel meer zou helpen in de verlaging van de druk, waaronder de administratieve druk, is een verandering van cultuur in de zorgmarkt. Een cultuur met meer vertrouwen in elkaars professionaliteit, een cultuur waarin buiten traditionele zorgsilo’s effectief wordt samengewerkt en een cultuur waarin incentives niet voornamelijk worden bepaald door budgetten, maar door een gemeenschappelijk streven naar verbeterde gezondheid en kleinere gezondheidsverschillen.
In de tussentijd vind ik het zeer verstandig om door te gaan met innovatief werk, om te onderzoeken of en hoe moderne technologie ingezet kan worden als assistent van de zorgprofessional. Het is echter véél te vroeg om nú al effecten hiervan te verwachten die de zorgmarkt verlichten.